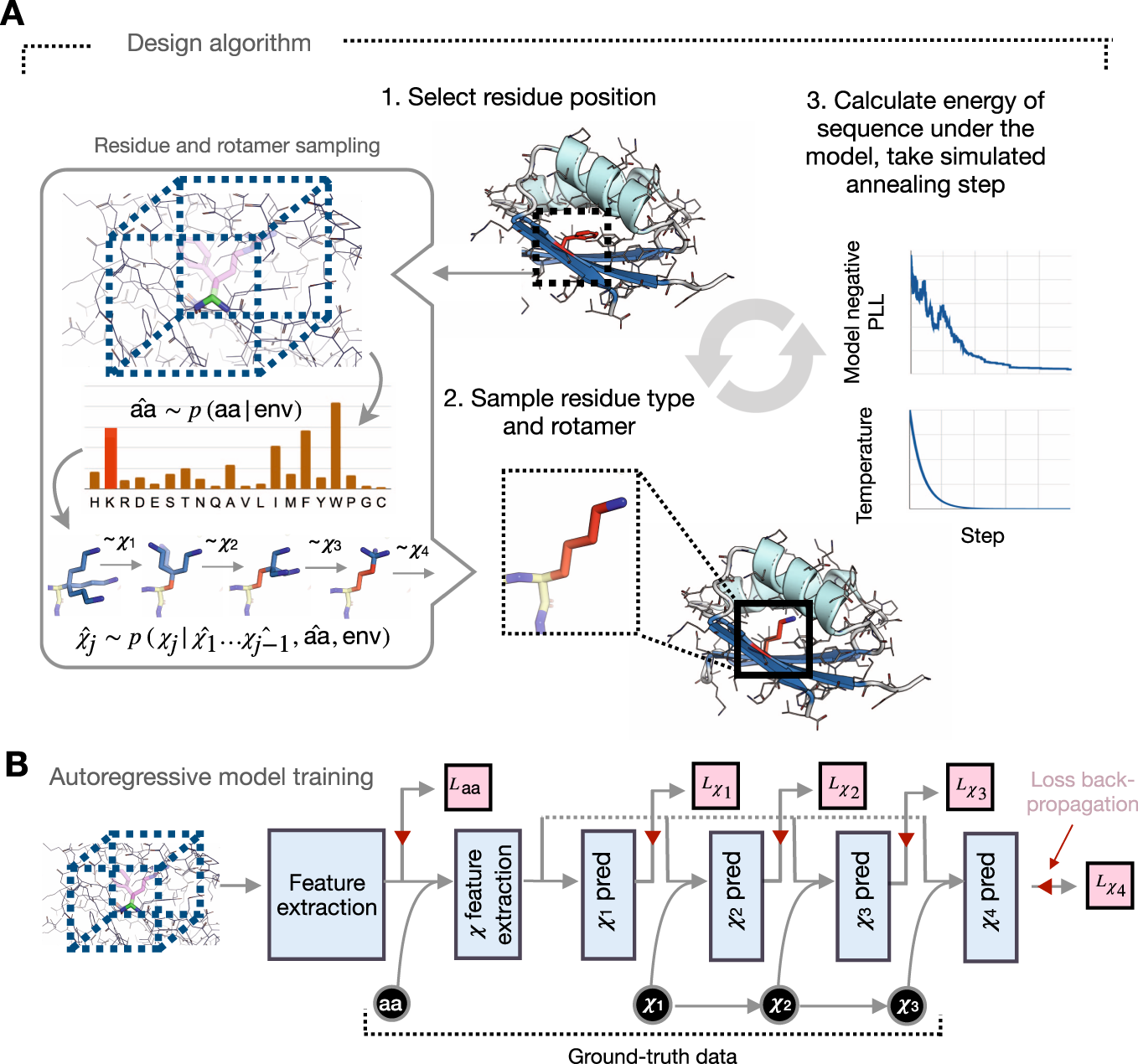
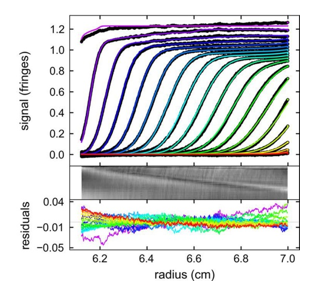
Molecules | Free Full-Text | Evaluation of Peptide/Protein Self-Assembly and Aggregation by Spectroscopic Methods
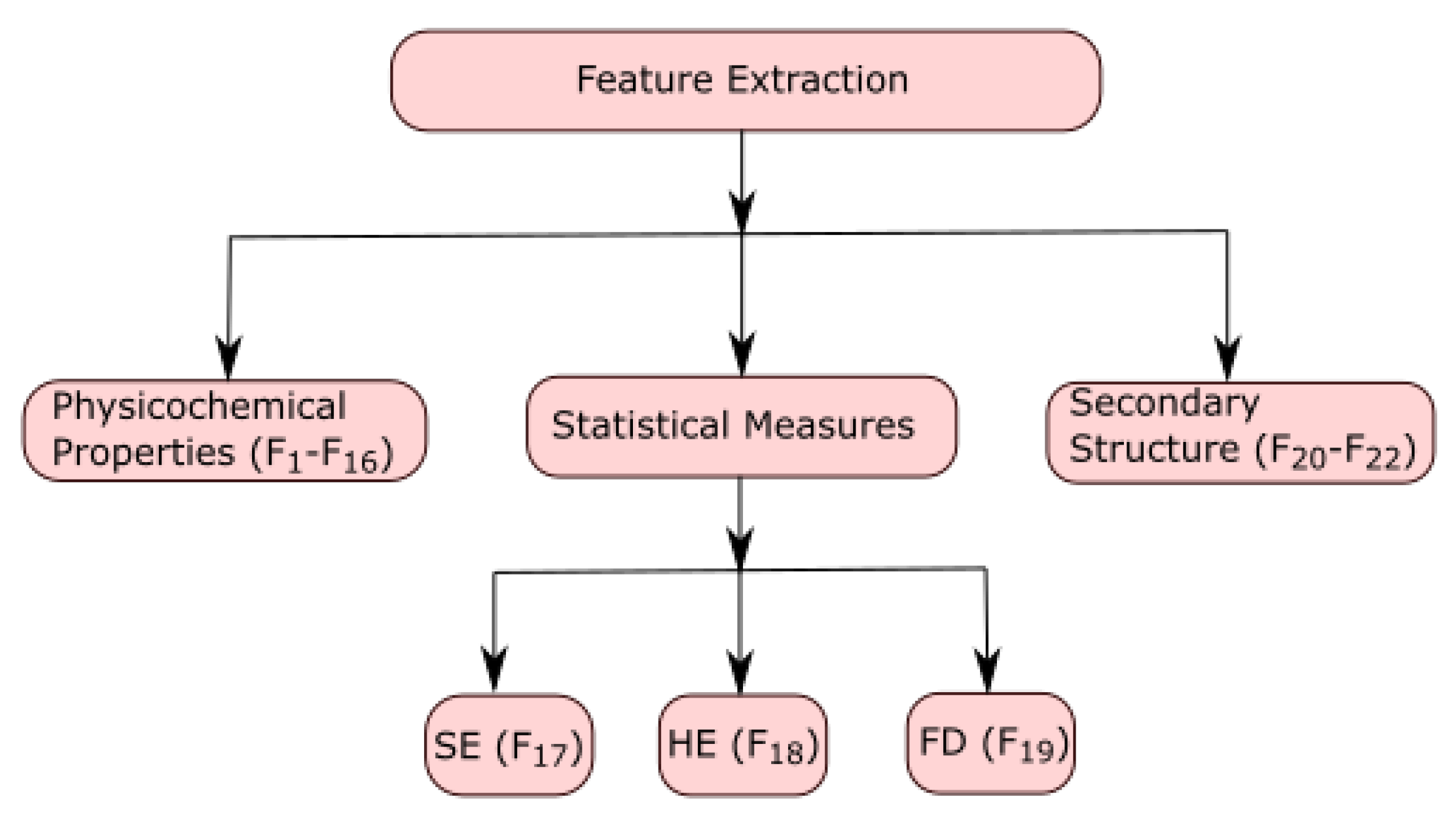
Mathematics | Free Full-Text | Unsupervised Learning for Feature Representation Using Spatial Distribution of Amino Acids in Aldehyde Dehydrogenase (ALDH2) Protein Sequences

A method to identify and quantify the complete peptide composition in protein hydrolysates - ScienceDirect
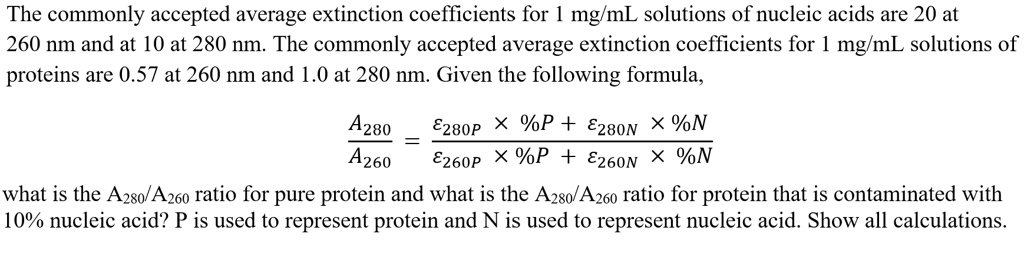
SOLVED: The commonly accepted average extinction coefficients for mg/mL solutions of nucleic acids are 20 at 260 nm and at 10 at 280 nm The commonly accepted average extinction coefficients for mglmL

Impact of external amino acids on fluorescent protein chromophore biosynthesis revealed by molecular dynamics and mutagenesis studies - ScienceDirect

SOLVED: It is possible to estimate the molar extinction coefficient of a protein from knowledge of its amino acid composition, as shown from your experiences with ExPASY: From the molar extinction coefficient
Complex folding and misfolding effects of deer-specific amino acid substitutions in the β2-α2 loop of murine prion protein | Scientific Reports
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/4-Table3-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar

Molar absorption coefficients for Trp, Tyr, and cystine based on an... | Download Scientific Diagram
Engrailed and Hox homeodomain proteins contain a related Pbx interaction motif that recognizes a common structure present in Pbx. - Abstract - Europe PMC

Observed and predicted molar absorption coefficients at 280 nm for 80... | Download Scientific Diagram
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/3-Table2-1.png)
PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar
![PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar PDF] Calculation of protein extinction coefficients from amino acid sequence data. | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/af80a5d2beee749cff2f5d98dc4a8f0dfdd48074/2-Table1-1.png)