Hidden Plant Stem Cells Could Hold the Key to Feeding the Future
Plant scientists discovered hidden stem cell regulators tied to growth and crop size. Their breakthrough could transform how we grow food, fuel, and resilient harvests.
Plant stem cells play a vital role in producing the world’s food, livestock feed, and renewable fuels. They are the foundation of plant growth, yet many aspects of how they work remain a mystery. Past studies have struggled to identify several of the key genes that govern stem cell activity.
Mapping the Genetic Regulators of Growth
For the first time, researchers at Cold Spring Harbor Laboratory (CSHL) have charted two well-known stem cell regulators across thousands of individual maize and Arabidopsis shoot cells. In the process, they also uncovered previously unknown regulators in both plants and connected some of them to size differences in maize. The technique they developed to recover rare stem cells could be applied broadly across many plant species.
CSHL Professor David Jackson explains: “Ideally, we would like to know how to make a stem cell. It would enable us to regenerate plants better. It would allow us to understand plant diversity. One thing people are very excited about is breeding new crops that are more resilient or more productive. We don’t yet have a full list of regulators—the genes we need to do that.”
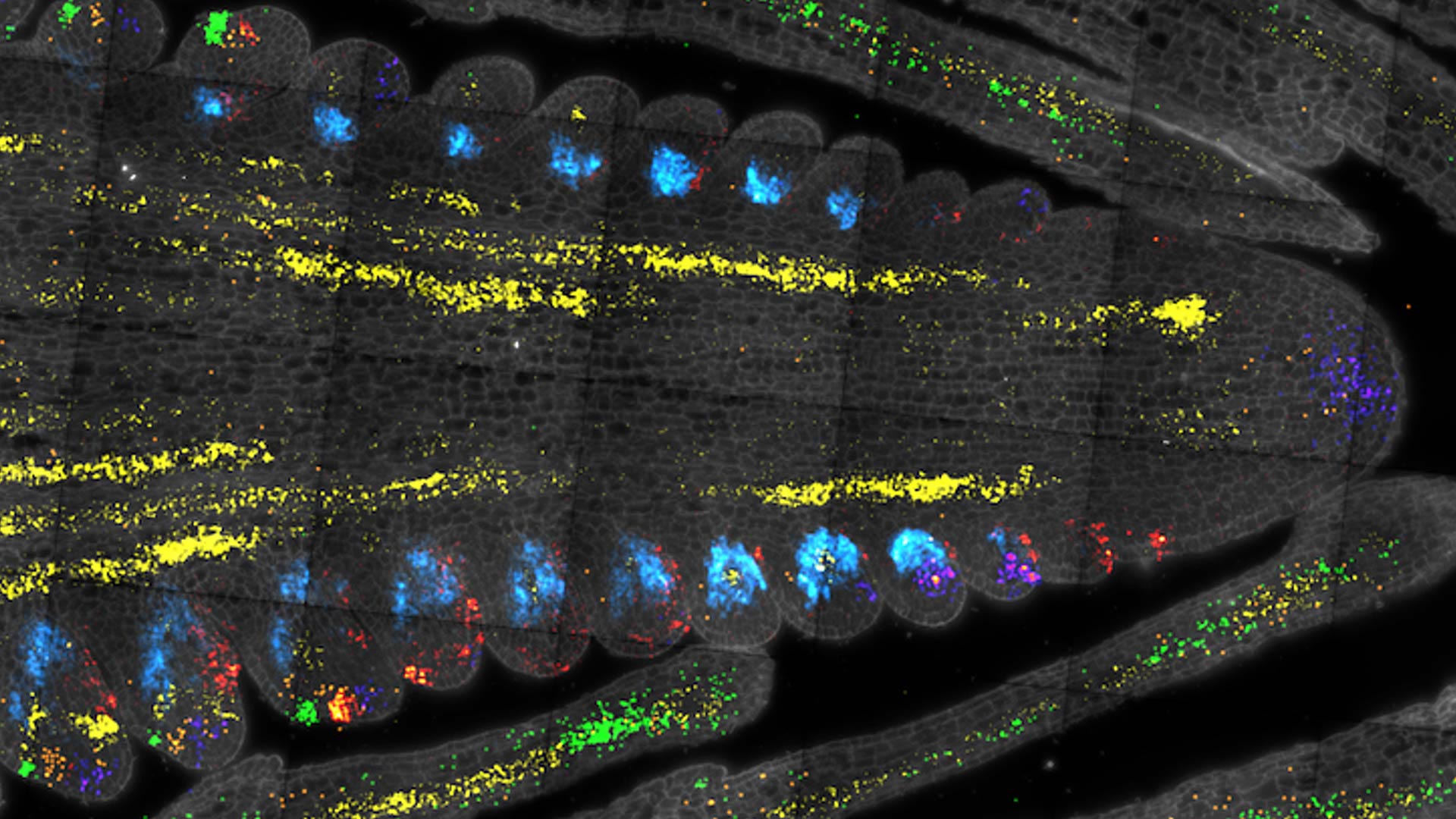
Jackson’s team began by investigating two widely studied regulators, CLAVATA3 and WUSCHEL. To access the stem cells, former postdoctoral researcher Xiaosa Xu carefully removed small sections of maize and Arabidopsis shoots. The researchers then turned to a “microfluidics” device to separate the cells one by one, convert their RNA into DNA, and attach a tag showing the identity of each cell.
Building a Gene Expression Atlas for All
The process, called single-cell RNA sequencing, allows researchers to see how genes are expressed in thousands of cells at once. “The great thing is that you have this atlas of gene expression,” Jackson says. “When we publish that, the whole community can use it. Other people interested in maize or Arabidopsis stem cells don’t have to repeat the experiment. They will be able to use our data.”
Single-cell RNA sequencing allowed the team to recover about 5,000 CLAVATA3 and 1,000 WUSCHEL-expressing cells. Next, they identified hundreds of genes that were preferentially expressed in both maize and Arabidopsis stem cells, suggesting they may be evolutionarily important across many plant species. From there, they were able to link certain stem cell regulators to productivity in maize. Such links could someday help breeders select specific strains for food, animal feed, or fuel production.
A Foundation for the Future of Agriculture
“It’s foundational knowledge that could guide research for the next decade,” Jackson says. “It can be used not only by developmental biologists, but physiologists, who think about how corn ears grow and how to improve productivity, and then breeders.”
Reference: “Large-scale single-cell profiling of stem cells identifies redundant regulators of shoot development and yield trait variation” by Xiaosa Xu, Michael Passalacqua, Brian Rice, Edgar Demesa-Arevalo, Mikiko Kojima, Yumiko Takebayashi, Xingyao Yu, Benjamin Harris, Yuchen Liu, Andrea Gallavotti, Hitoshi Sakakibara, Jesse Gillis and David Jackson, 26 August 2025, Developmental Cell.
DOI: 10.1016/j.devcel.2025.07.024
Never miss a breakthrough: Join the SciTechDaily newsletter.
Source link

